Documentation > Documentation
You should see something like below. The documentation concerning the GUI is provided within the GUI, some basic information can also be found here.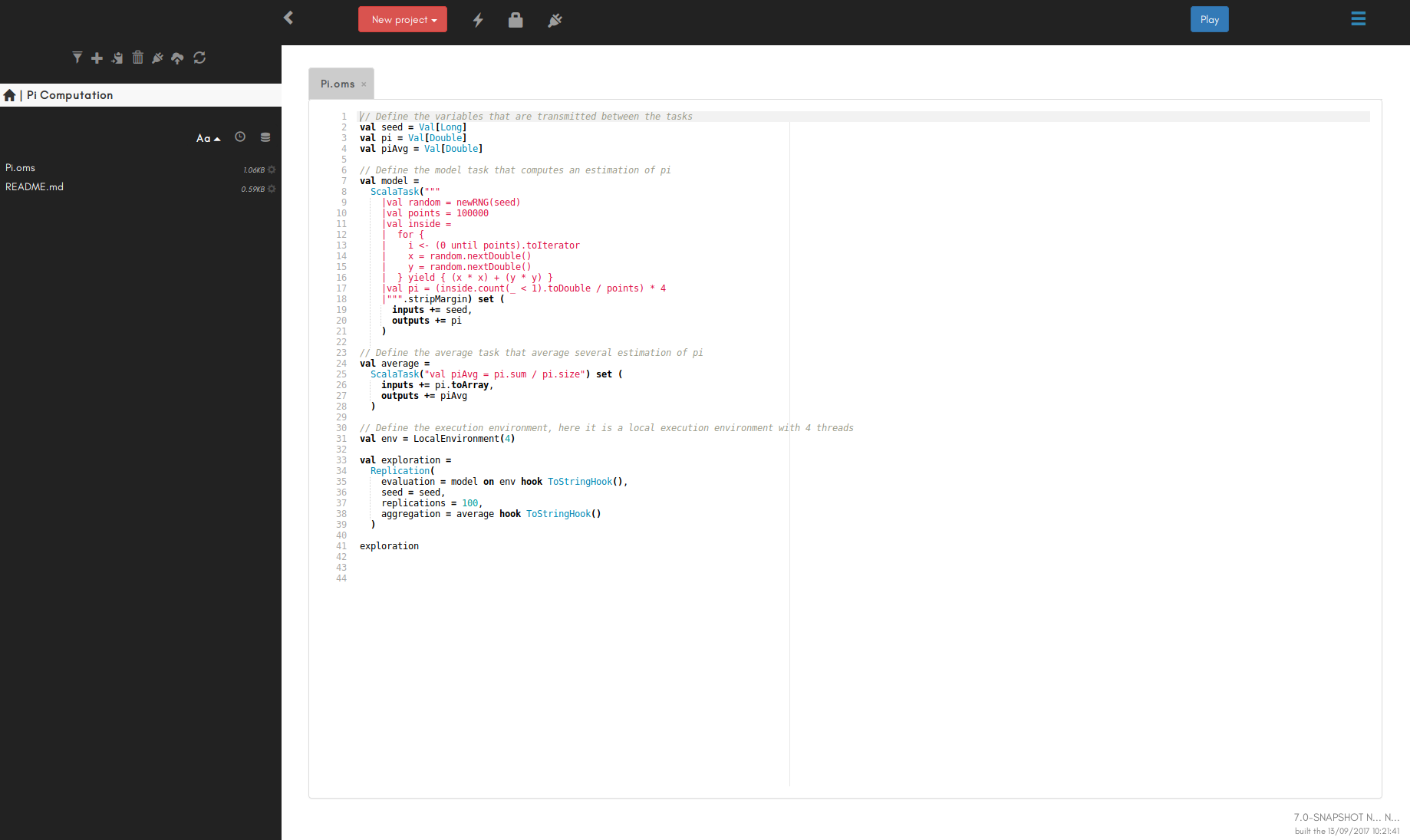
In that case, OpenMOLE still asks for your previous cyphering password. To provide it at launch time, use the
A console workflow is launched like this:
Content:
Running OpenMOLE with a Graphical User Interface 🔗
The default way to run the OpenMOLE application is with a graphical user interface (GUI). To do so, just execute theopenmole file in the folder you downloaded, it will bring up the application in your web browser.
OpenMOLE supports Chrome and Firefox, so if you are using another default web browser please copy-paste the OpenMOLE url http://localhost:[port] in one of these browsers.
You should see something like below. The documentation concerning the GUI is provided within the GUI, some basic information can also be found here.

GUI on a remote machine 🔗
To run OpenMOLE on a remote machine you should execute the following commandopenmole --remote --port portnumber.
The first time you launch it, you will be prompted to choose a password.
Then you can remotely connect to OpenMOLE using the url https://remotemachine:portnumber (note that the "https://" part is important).
When you connect to OpenMOLE through your remote, you will be asked for the password you chose previously.
Running OpenMOLE in headless mode 🔗
OpenMOLE offers a headless mode for running scripts. You can enable it thanks to the-s option: ./openmole -s /path/to/you/mole/script.
In that case, OpenMOLE still asks for your previous cyphering password. To provide it at launch time, use the
-pw option: ./openmole -s /path/to/your/mole/script --password password.
A better practice is to write this password in a file readable by OpenMOLE only, and use ./openmole -s /path/to/your/mole/script --password-file password.txt.
Interactive console mode 🔗
OpenMOLE also offers an interactive console mode. To launch the console executeopenmole -c in a console.
The only differences between the scripts in the console mode and the ones from the editor in the GUI are the ways you launch the execution of a workflow, you cancel it, and you follow the execution progress.
A console workflow is launched like this:
val exploration =
DirectSampling(
evaluation = myModel on env,
sampling = mySampling
)
val ex = exploration start
ex and the env variables created above, you can follow the progress of the execution by using the commands print(ex) and print(env).
To cancel the execution you should use ex.cancel.
Launching options 🔗
OpenMOLE comes with several launching options. Executeopenmole -h in a terminal to list them all.