Documentation > Plug
OpenMOLE provides a NetLogo task which expects the following parameters:
The
Similarly, an output of the model is considered and collected by OpenMOLE at the end of each model execution. It is written as
The arguments for a NetLogoTask are the following :
It goes the same for all additional source files (
Since the Fire model is stochastic, we are interested in doing replications for each instance of the
When designing your experiment, you will have to find a compromise between the precision on stochasticity and the number of parameter points explored. More elaborated methods, in the case of a calibration of a stochastic model with a genetic algorithm for example, will automatically deal with this compromise (see this page for more info on genetic algorithms and calibration).
You can get the NetLogo implementation of the model here.
NetLogo is a graphical framework. As such, many variables of a model developed in NetLogo are set through widgets (a graphical component). In the NetLogo World, setting or getting a value on the model inputs is generally achieved by calling set or get on the widget object. In OpenMOLE however, the NetLogo program has to be parameterised without the GUI. Models must be used in headless mode with OpenMOLE. This is not a problem because globals with unspecified values in the OpenMOLE
There is no strict requirement for a normal model to run, except that the user cannot access plots nor reporters, or export images of the world. Global variables defined in the GUI (as sliders or switches for example) are still available and can be set from the OpenMOLE script.
We recommend however the following ``best practices'':
These best practices may be too constraining for simple experiments, we give therefore the following less strict guidelines:
The following shows the application of these guidelines to the NetLogo model introduced before.

This excerpt shows the

We propose here a simple method to better organize your code in order to make it manipulable by OpenMOLE:
Content:
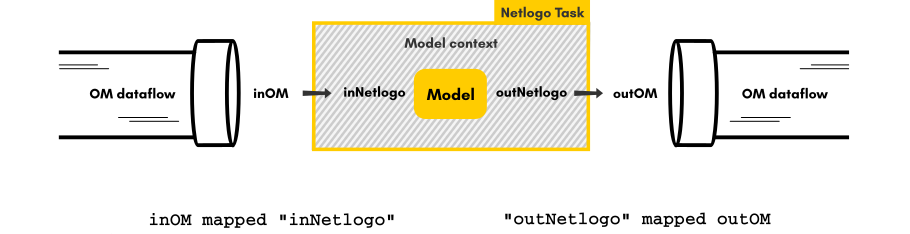
NetLogo Task 🔗
NetLogo is a widely used agent-based modeling platform and language developed by CCL at Northwestern University (see the official website). It is conceived to be accessible to non-programmers and thus enhances inter-disciplinarity in the construction of simulation models, but can also be used to program large scale complex models (despite its bad reputation, see this paper testing NetLogo speed performance and suggesting code improvements). As it runs on the JVM, it is naturally integrated into OpenMOLE.OpenMOLE provides a NetLogo task which expects the following parameters:
- the path to the NetLogo model, i.e. the
.nlogosource file, - the list of NetLogo commands to be run by OpenMOLE.
NetLogo5Task, NetLogo6Task and NetLogo7Task, be sure to select the right version of the task according to the version of NetLogo you are using (5, 6 or 7).
Here is an example on how to write the NetLogo task for the model Fire.nlogo:
val cmds = List(
"random-seed ${seed}",
"setup",
"while [any? turtles] [go]")
val fireTask =
NetLogo7Task(workDirectory / "Fire.nlogo", cmds) set (
inputs += seed,
inputs += density mapped "density",
outputs += (density, seed),
outputs += burned mapped "burned-trees"
)random-seedinitializing the random number generator of NetLogo using the seed provided by OpenMOLE,setupcalling the setup function of the nlogo file,gorunning the model. For this particular model, this function is called until no more turtles are active.
go in the NetLogo GUI don't forget that you will need something like while [condition] [go] in the command list, as openmole won't loop your go function by default.
The
replication and density OpenMOLE variables are used as parameters of the NetLogo program.
Therefore they appear as inputs of the NetLogoTask.
Similarly, an output of the model is considered and collected by OpenMOLE at the end of each model execution. It is written as
netLogoOutputs in the definition of the task.

The arguments for a NetLogoTask are the following :
scriptNetLogo model (File), mandatorylaunchingCommandsNetLogo commands to be executed, mandatoryembedWorkspaceshould the workspace be embedded for execution of the model (use if you have source files or extensions in the model directory), optional, defaults to falsereuseWorkspaceshould the same workspace be reused when executing on a given jvm (use to avoid MetaSpace errors with large NetLogo models loading several extensions when executed a large number of times on the same jvm), optional, defaults to falseseedrandom seed, optional, defaults to None
set :
inputs/outputssimilar as for any task- mapped input: the syntax
inputs += prototype mapped "netlogo-variable"establishes a link between the workflow variableprototype(Val) and the corresponding netlogo variable namenetlogo-variable(String). If the variables have the same name, you can use the shorter syntaxinputs += prototype.mapped - mapped output: similar syntax to collect outputs of the model (the string can be any NetLogo command)
netLogoInputs and netLogoOutputs is deprecated, but still works until further notice, for compatibility reasons.
Embedding NetLogo extensions and sources 🔗
Several NetLogo models rely on extensions (which are basically jars providing new primitives in interaction with the NetLogo workspace). By default, NetLogo will search extensions in its installation folder (which will not work here, as OpenMOLE embeds NetLogo as a jar), but also in the model directory. To be able to run a model with extensions, put the extension folder in the same directory as the model source, and activate the optionembedWorkspace.
It goes the same for all additional source files (
.nls generally used for large models) or configuration files.
An example of NetLogo model exploration 🔗
We now present step by step how to explore a NetLogo model. The Fire model is a common NetLogo example available in the NetLogo common model library. It studies the percolation of a fire in a forest depending on the density of the forest. This model has one input:density, and one output: percent-burned.

The simulation 🔗
We would like to study the impact of thedensity factor for a fixed population size.
To do this, let's build a design of experiment where the density factor ranges from 20% to 80% by steps of 10.
Since the Fire model is stochastic, we are interested in doing replications for each instance of the
density factor.
Results for each replication will be stored it in a CSV file.
In this example case, we will perform 10 replications per step.
This stays a too small sample to draw up any robust conclusion on this simple model, but we take this value here for the sake of illustration.
When designing your experiment, you will have to find a compromise between the precision on stochasticity and the number of parameter points explored. More elaborated methods, in the case of a calibration of a stochastic model with a genetic algorithm for example, will automatically deal with this compromise (see this page for more info on genetic algorithms and calibration).
You can get the NetLogo implementation of the model here.
The Design of Experiment 🔗
We first need to define two OpenMOLE variables in order to repeat our experience 10 times for every step of thedensity exploration.
These two variables are:
- an integer (Int) representing the seed of the random number generator for exploring the replications,
- a Double to set the value of
density
val density = Val[Double]
val seed = Val[Int]
val burned = Val[Double]val sampling =
(density in (20.0 to 80.0 by 10.0)) x
(seed in RandomSequence[Int](size = 10))- 10 replications with 10 different seeds for
density= 20% - 10 replications with 10 different seeds for
density= 30% - ...
- 10 replications with 10 different seeds for
density= 80%
Storing the results 🔗
OpenMOLE usually delegates the tasks execution to many different computers. To gather the results of these remote executions, we use a mechanism called hooks. Hooks can be assimilated to a listener that saves or display results. Most of the time it is enough to use the hook keyword provided to either display or store the results of the exploration process. Hooks are more thoroughly described in a specific section of the documentation.Bringing all the pieces together 🔗
Now that we have defined each component, we can compose the workflow that brings all the pieces of the simulation together:val density = Val[Double]
val seed = Val[Int]
val burned = Val[Double]
val sampling =
(density in (20.0 to 80.0 by 10.0)) x
(seed in RandomSequence[Int](size = 10))
val cmds = List(
"random-seed ${seed}",
"setup",
"while [any? turtles] [go]")
val fireTask =
NetLogo7Task(workDirectory / "Fire.nlogo", cmds) set (
inputs += seed,
inputs += density mapped "density",
outputs += (density, seed),
outputs += burned mapped "burned-trees"
)
DirectSampling(
evaluation = fireTask,
sampling = sampling
) hook (workDirectory / "result")Import wizard 🔗
When working with models with a large number of parameters and/or outputs, writing the script can be already painful just when defining the prototypes. Fortunately, OpenMOLE includes an import wizard in its interface, for several languages including NetLogo. To use it, use the buttonNew project and the option Import your model.
The wizard will detect your parameters and outputs (defined in the GUI), the resources used (such as external files), and propose in an interactive window to change these.
Once you validate, a minimal script is created.
Headless NetLogo model 🔗
Netlogo combines a GUI created by the user, in which they can define parameters and run functions, and the source code itself. Sometimes, you have to modify your code slightly to make it purely headless so that it can be run everywhere.NetLogo is a graphical framework. As such, many variables of a model developed in NetLogo are set through widgets (a graphical component). In the NetLogo World, setting or getting a value on the model inputs is generally achieved by calling set or get on the widget object. In OpenMOLE however, the NetLogo program has to be parameterised without the GUI. Models must be used in headless mode with OpenMOLE. This is not a problem because globals with unspecified values in the OpenMOLE
NetLogoTask will take the default values defined in widgets.
There is no strict requirement for a normal model to run, except that the user cannot access plots nor reporters, or export images of the world. Global variables defined in the GUI (as sliders or switches for example) are still available and can be set from the OpenMOLE script.
We recommend however the following ``best practices'':
- Do not keep any global variable in the GUI, so that reading the code of the headless model explicitly gives all globals
- To set variables for the experiment, one can either use input mapping or string substitution within a NetLogo command (a typical example being
setup-experiment var1 var2and a specific setup procedure for experiments. This allows to know explicitly variables concerned by the experiments, and not forget to setup any global. This procedure can be used upstream the actual setup, acting as a setup of parameters in the GUI (the actual setup will need then to test if current mode is headless before clearing-all for example) - The use of direct variable setting is particularly useful when dealing with large arrays that will fail with string substitution.
When using this way of setting the variables, the user must be careful of not calling the
clear-allprocedure at the beginning of his code (in practice, globals are set before the call to user commands - aclear-allis done before that, to ensure workspace is clean in the case of pooling) - Keep two files of the model, one for the standard model and one for the headless. This imply having all the code within
.nlsinclude files (except globals, breed and variables declarations). A good way to proceed is to separate procedures by types of agents and/or purpose.
These best practices may be too constraining for simple experiments, we give therefore the following less strict guidelines:
- Limit your usage of widget implicit globals. We prefer explicit globals using the globals primitive, because you need to re-define all the important values from your program in OpenMOLE before you launch it on a remote computing environment.
- Do not use the same functions
setupandgoto setup your program on a remote environment. On the remote environment, the NetLogo program is initialised and launched only once, so there is no need to call aclear-allprimitive with each setup. When running on distributed environments,clear-allis not your best friend.clear-allerases all the globals passed by OpenMOLE to your program before it starts and will make it crash.} - Do not directly use the implicit globals created by means of a widget. Although you can you can access and overwrite implicit globals in OpenMOLE, it prevents OpenMOLE from mapping explicitly its prototypes to the NetLogo globals.
The following shows the application of these guidelines to the NetLogo model introduced before.
Application to the Fire model 🔗
Fire.nlogo has a widget slider nameddensity which is a global implicit.

This excerpt shows the
initial-trees and burned-trees variables which are explicit globals.
They can be used directly in OpenMOLE.

We propose here a simple method to better organize your code in order to make it manipulable by OpenMOLE:
-
First we do not use the implicit globals, so we create an explicit global variable,
myDensity, corresponding to the implicit one (density) :

-
Second, we use this new variable in the setup procedure (it replaces the former):

And we update the explicit variable with the former implicit density variable
At this moment, your program does not work any more in NetLogo, it's normal, don’t panic :). -
Third, we call this function in our setup function, after the
clear-allprimitives.
-
Now, the program works in NetLogo’s graphical mode.
We still need to create another setup function without the call to
clear-alland toinit-globals. Remember that these two calls don't cope well with distributed executions.